Publications
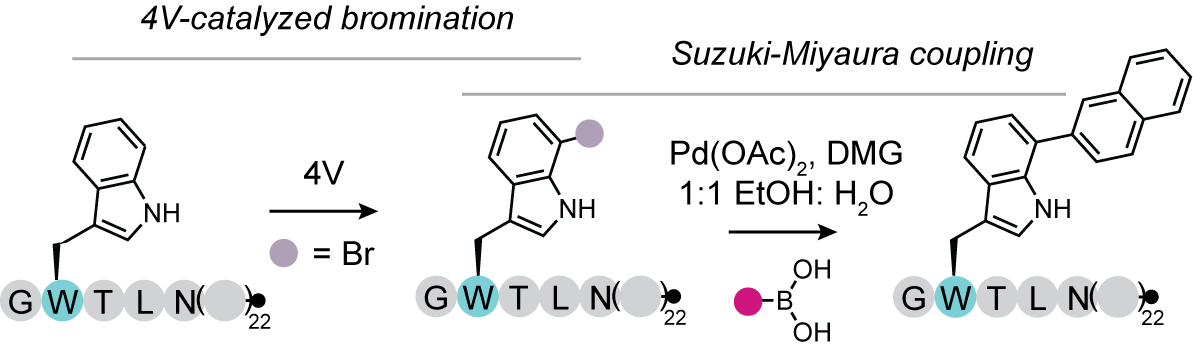
27. Bridge, H.N., Radziej, C.L., Weeks, A.M. Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling. bioRxiv 2025 (link).
26. Xu, Z., Mazurkiewicz, L.E., Olivetta, M., Morrissey, M., Dudin, O., Weeks, A.M., Coyle, S.M. A self-organizing single-cell morphology circuit optimizes Podophrya collini predatory trap structure. bioRxiv 2025 (link).
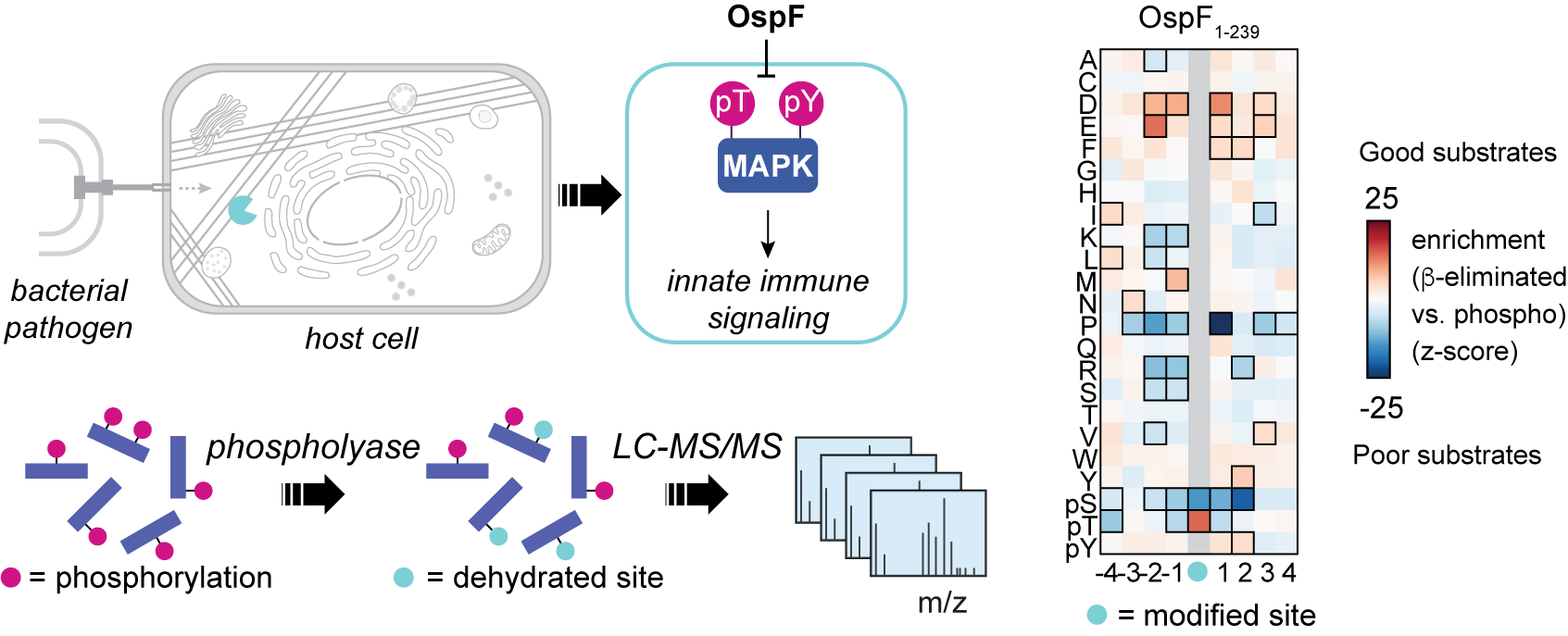
25. Radziwon, K., Campbell, L.A., Mazurkiewicz, L.E., Jalalishvili, S., Eppinger, I., Parikh, A., and Weeks, A.M. Phosphoproteome-derived peptide libraries for deep specificity profiling of phosphatases and phospholyases. bioRxiv 2025 (link).
23. Weeks, A.M. PEARLs of wisdom for ribosome-independent peptide bond synthesis. Proc. Natl. Acad. Sci. U. S. A. 2025, 122, e2504930122. (Commentary) (link)
22. Schwarz, E.M., Noon, J.B., Chicca, J.D., Garceau, C., Li, H., Antoschechkin, I., Ilík, V., Pafčo, B., Weeks, A.M., Homan, E.J., Ostroff, G.R., Aroian, R.V. Hookworm genes encoding intestinal excreted-secreted proteins are transcriptionally upregulated in response to the host’s immune system. bioRxiv 2025 (link).
21. Chung, Y., Yim, C., Pereira, G., Son, S., Kjølbye, L.R., Mazurkiewicz, L.E. and Weeks, A.M., Förster, F., von Heijne, G., de Souza, P.C.T., and Kim, H. Spc2 modulates substrate- and cleavage site-selection in the yeast signal peptidase complex. J. Cell Biol. 2024, 223, e202211035 (link).
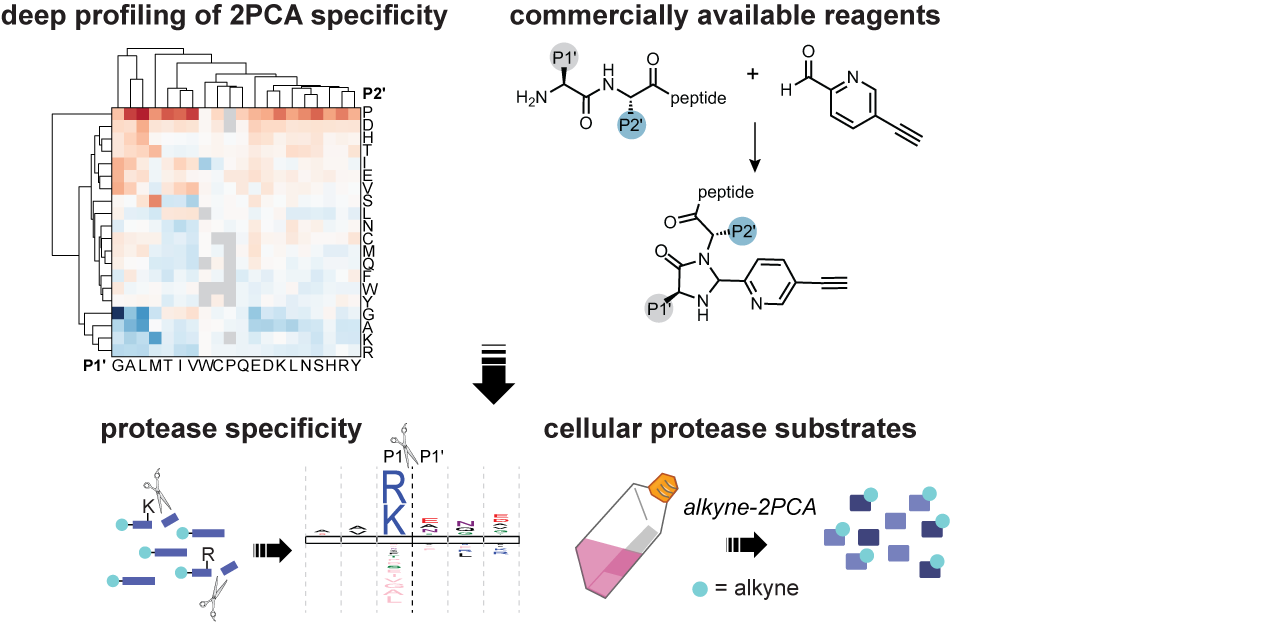
20. Bridge, H.N., Leiter, W., Frazier, C. L., and Weeks, A. M. An N terminomics toolbox combining 2-pyridinecarboxaldehyde probes and click chemistry for profiling protease specificity. Cell Chemical Biology 2024, 31, 534-549.e8. (link).
Preprint: bioRxiv 2023 (link)
19. Schaefer, K., Lui, I., Byrnes, J.R., Kang, E., Zhou, J., Weeks, A.M., and Wells, J.A. Direct identification of proteolytic cleavages on living cells using a glycan-tethered peptide ligase. ACS Central Science 2022, 8, 1447-1456. (link)
18. Amiridis, A.A. and Weeks, A.M. Mapping cell surface proteolysis with plasma membrane-targeted subtiligase. Methods Mol. Biol. 2022, 2456, 71-83. (Invited methods paper) (link)
17. Byrnes, J.R., Weeks, A.M., Shifrut, E., Carnevale, J., Kirkemo, L., Ashworth, A., Marson, A., and Wells, J.A. Hypoxia is a dominant remodeler of the effector T cell surface proteome relative to activation and regulatory T cell suppression. Mol. Cell. Proteomics 2022, 21, 100217. (link)
16. Weeks, A.M. Kinases leave their mark on caspase substrates. Biochem. J., 2021, 478, 3179-3184. (Invited commentary) (link)
15. Weeks, A.M. Spatially resolved tagging of proteolytic neo-N termini with subtiligase-TM. J. Membrane Biol., 2021, 254, 119-125. (Invited ‘Up-and-Coming Scientist’ review) (link)
14. Weeks, A.M., Byrnes, J.R., Lui, I., and Wells, J.A. Mapping proteolytic neo-N termini at the surface of living cells. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2018809118. (link) (preprint)
13. Radziwon, K. and Weeks, A.M. Protein engineering for selective proteomics. Curr. Opin. Chem. Biol. 2021, 60, 10-19. (Review) (link) (pdf)
12. Frazier, C.L. and Weeks, A.M. Engineered peptide ligases for cell signaling and bioconjugation. Biochem. Soc. Trans. 2020, 48, 1153-1165. (Review) (link) (pdf)
11. Weeks, A.M.† and Wells, J.A.† N-terminal modification of proteins with subtiligase specificity variants. Curr. Protoc. Chem. Biol. 2020, 12, e79. †corresponding author. (Invited methods paper) (link) (pdf)
10. Weeks, A.M.† and Wells, J.A.† Subtiligase-catalyzed peptide ligation. Chem. Rev. 2020, 120, 3127-3160. †corresponding author. (Review) (link) (pdf)
9. Weeks, A.M.*, Wang, N.*, Pelton, J.G., and Chang, M.C.Y. Entropy drives selective fluorine recognition in the fluoroacetyl-CoA thioesterase from Streptomyces cattleya. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E2193-E2201. *equal contribution.
8. Weeks, A.M. and Wells, J.A. Engineering peptide ligase specificity by proteomic identification of ligation sites. Nat. Chem. Biol. 2018, 14, 50-57.
7. Lin, S.*, Yang, X.*, Jia, S., Weeks, A.M., Lee, P.S., Hornsby, M., Nichiporuk, R.V., Iavarone, A.T., Wells, J.A., Toste, F.D.†, and Chang, C.J.† Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355, 597-602. *equal contribution. †corresponding author.
6. Weeks, A.M., Wadoux, R.D.P., Keddie, N.S., O’Hagan, D., and Chang, M.C.Y. Molecular recognition of fluorine impacts substrate selectivity in the fluoroacetyl-CoA thioesterase FlK. Biochemistry 2014, 53, 2053-2063.
5. Weeks, A.M. and Chang, M.C.Y. Catalytic control of enzymatic fluorine specificity. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 19667-19672.
4. Bond-Watts, B.B., Weeks, A.M., and Chang, M.C.Y. Biochemical and structural characterization of trans-enoyl-CoA reductase from Treponema denticola. Biochemistry 2012, 51, 6827-6837.
3. Walker, M.C., Wen, M., Weeks, A.M., Chang, M.C.Y. Temporal and fluoride control of secondary metabolism regulates cellular organofluorine biosynthesis. ACS Chem. Biol. 2012, 7, 1576-1585.
2. Weeks, A.M. and Chang, M.C.Y. Constructing de novo biosynthetic pathways for chemical synthesis inside living cells. Biochemistry 2011, 50, 5404-5418. (Review)
1. Weeks, A.M., Coyle, S.M., Jinek, M., Doudna, J.A., and Chang, M.C.Y. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry 2010, 49, 9269-9279.